Proteins at interfaces
Atomically detailed insights into their structure, dynamics and bio-activity
Proteins constitute the largest and most widely employed class of biomolecules in surface functionalization. There is a wide range of applications in biotechnology that rely on protein adsorption such as biocompatible interfaces, biosensorifis, and interfaces for tissue engineering and regenerative medicine. One common requirement in these applications is that the protein must remain bioactive upon deposition. Protein adsorption also represents an interesting and important fundamental problem because it is the result of a delicate interplay among protein–surface interactions, hydration forces, and the protein’s ability to change its conformation.
Fueled by its fundamental and technological interest, much effort has been devoted in understanding and controlling protein–surface interaction from an experimental point of view. Yet, the inherent difficulties to probe events at an atomistic level has limited our understanding of this complex process and as a result most devices dependent on protein-surface interactions are developed on a trial error approach. To bridge this gap and enable a knowledge based approach, over the past years we employed all-atom molecular dynamics simulations to study the adsorption of the most abundant plasma proteins to inorganic surfaces.
Our works have unraveled a wide range of surprising results, to name a few:
Our all-atom molecular dynamics simulations revealed vertical antibody adsorption to graphene, a critical feature for biosensors as it leaves the bioactive fragments accessible.
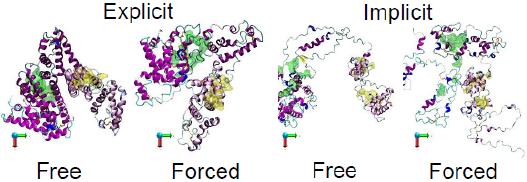
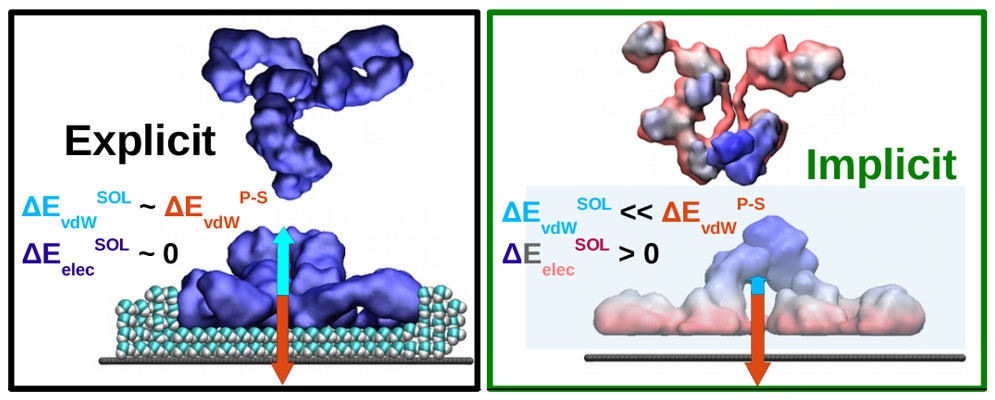
We discovered that water structure, particularly in the protein’s hydration layer, plays an active role in stabilizing protein structure during adsorption. Misrepresentation of this hydration layer, such as using implicit solvent methods, disrupts the energy balance and leads to systematic and spurious protein unfolding during adsorption.
We have shown that specific binding sites in the most abundant globulin proteins (IgG and Albumin) can also be used to program surface adsorption along well defined orientations.
Publications
M. Ortega, J. G. Vilhena, P. Rubio, P. A. Serena and R. Pérez. “Assessing the accuracy of implicit solvation models to describe the IgG adsorption over a hydrophobic surface“. Journal of Chemical Theory and Computation 15 (4), 2548 (2019).
P. Rubio, J. G. Vilhena, N. Takeuchi, P. A. Serena amd R Pérez. “Albumin (BSA) adsorption onto graphite stepped surfaces”. The Journal of Chemical Physics 146(21), 214704 (2017).

J. G. Vilhena, A. C. Dumitru, E. T. Herruzo, J. I. Mendieta, R. García, P. A. Serena and R. Pérez. “Adsorption orientations and immunological recognition of antibodies on graphene“. Nanoscale 8 (27), 13463 (2016).

J. G. Vilhena, P. Rubio, P. Vellosillo, P. A. Serena and R. Pérez. “Albumin (BSA) Adsorption over Graphene in Aqueous Environment: Influence of Orientation, Adsorption Protocol, and Solvent Treatment”. Langmuir 32 (7), 1742 (2016).