Mechanics of Biomolecules
From atomic workings of nucleic-acids up to protein’s nanomechanical maps
Biomolecular mechanics play a pivotal role in a myriad of biological processes, influencing everything from the replication and repair of genetic material to the interaction between various biomolecules inside the cell. By deciphering these mechanical properties, we aim to shed light on the inner workings of biological systems, offering physical insights essential for unlocking the mysteries of life itself.
A prime example of our focus lies in the study of double-stranded nucleic acids (DNA and RNA), where mechanical properties dictate crucial biological functions. From the localized movements of individual base pairs upon protein binding to the global folding of genome-length polymers, these biomolecules exhibit a broad spectrum of deformations across multiple length scales. Despite intensive research efforts, bridging the dynamics across these scales remains a formidable challenge.
In our pursuit, we’ve developed a simulation strategies that unveils the atomic underpinnings of nucleic acid mechanics, resolving long-standing paradoxes and offering profound insights. By synergistically combining simulation with experimental techniques such as Atomic Force Microscopy (AFM), Optical Tweezers (OT), and Magnetic Tweezers (MT), we connect our findings to processes occurring on larger scales. Some milestone achievements include:
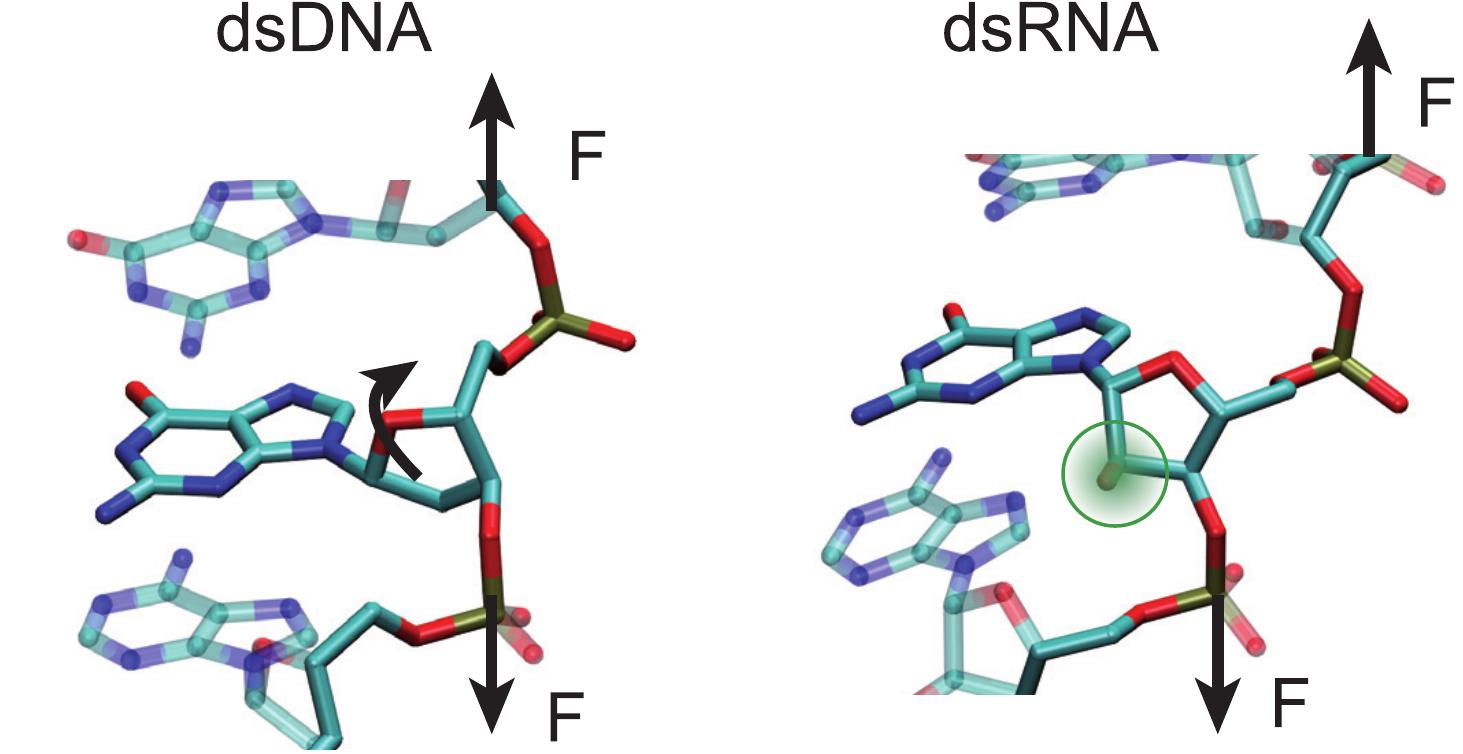
Resolving a decade-long paradox regarding the differential mechanical response of dsDNA and dsRNA. We’ve uncovered a physical mechanism explaining their counterintuitive twist-stretch behaviors, tracing back to their fundamental differences at the atomic level.
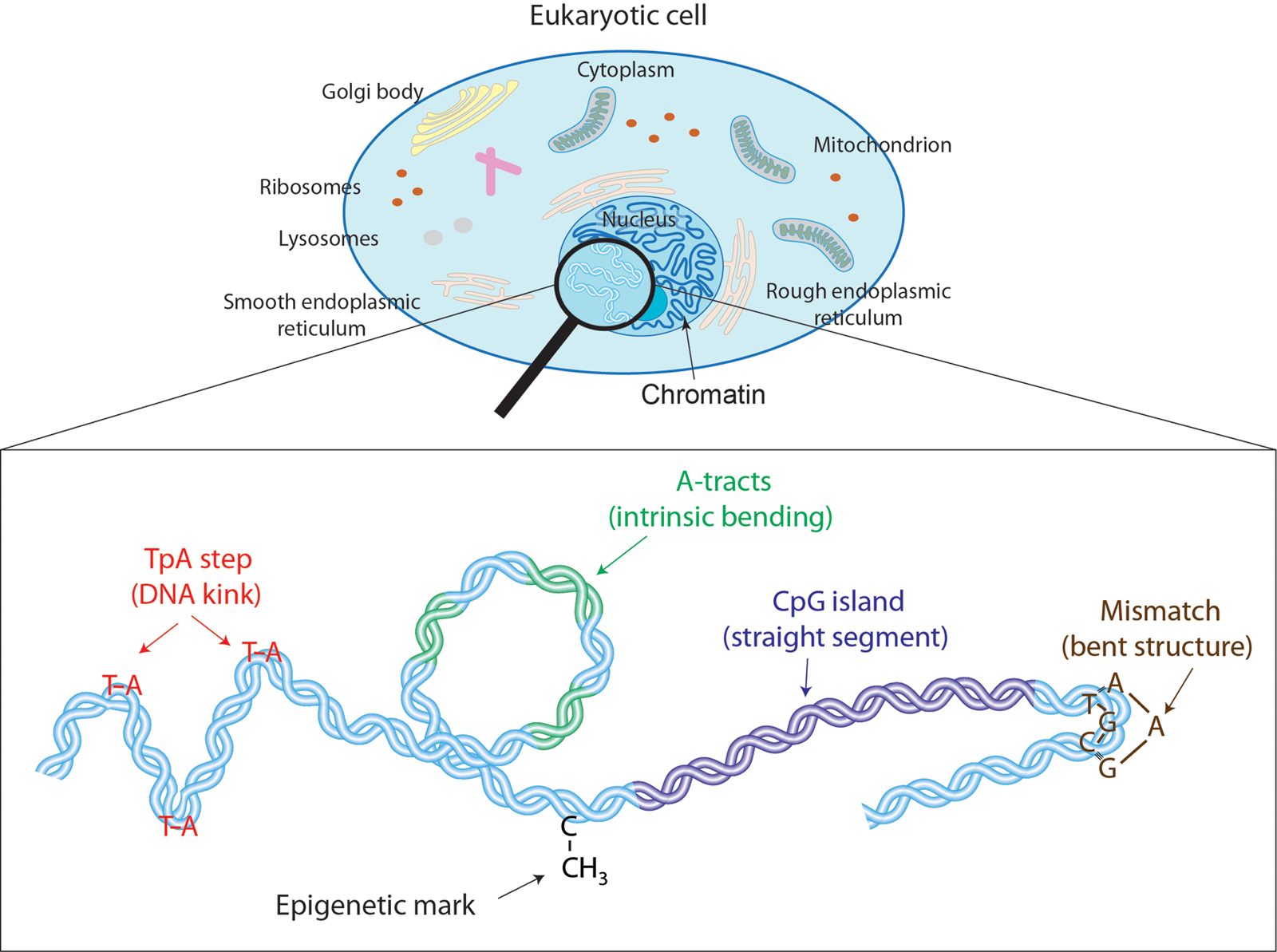
Demonstrating that DNA sequence acts not only as a chemical code but also as a physical one. By altering the sequence, we can significantly alter the molecule’s stiffness and correlate these changes with the activation or deactivation of various biological processes. Furthermore, we’ve elucidated how common mutations can reverse these behaviors, finely tuning the DNA’s mechanical and biological responses.
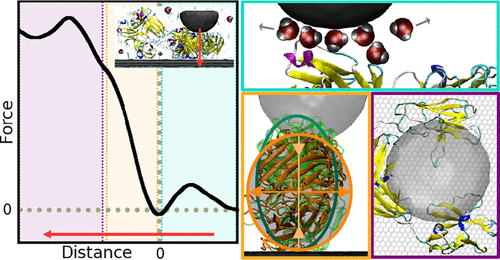
Conducting the first nano-mechanical mapping of a full-sized protein through atomically detailed simulations. This endeavor has not only elucidated the atomic deformations associated with previous Young modulus measurements but has also determined the technique’s spatial resolution limits. Additionally, we’ve uncovered exceptionally high lateral resolution in the non-contact regime, opening new avenues for exploration.
Publications
J. G. Vilhena, M. Ortega, M. Uhlig, R. García and R. Pérez. “A Practical Guide to Single Protein AFM Nanomechanical Spectroscopy Mapping: Insights and Pitfalls as Unraveled by All-Atom MD Simulations on Immunoglobulin G“. ACS Sensors 6(2), 553 (2021).
A. Marín, J. G. Vilhena, R. Pérez and F. Moreno. “A molecular view of DNA flexibility“. Quarterly Reviews of Biophysics 54, e8 (2021).
A. Marín, C. Aicart, Mikel. Marin, A. Martín, M. Suomalainen, A. Kannan, J. G. Vilhena, U. F. Greber, F. Moreno and R. Pérez. “Double-stranded RNA bending by AU-tract sequences“. Nucleic Acids Research 48 (22), 12917 (2020).
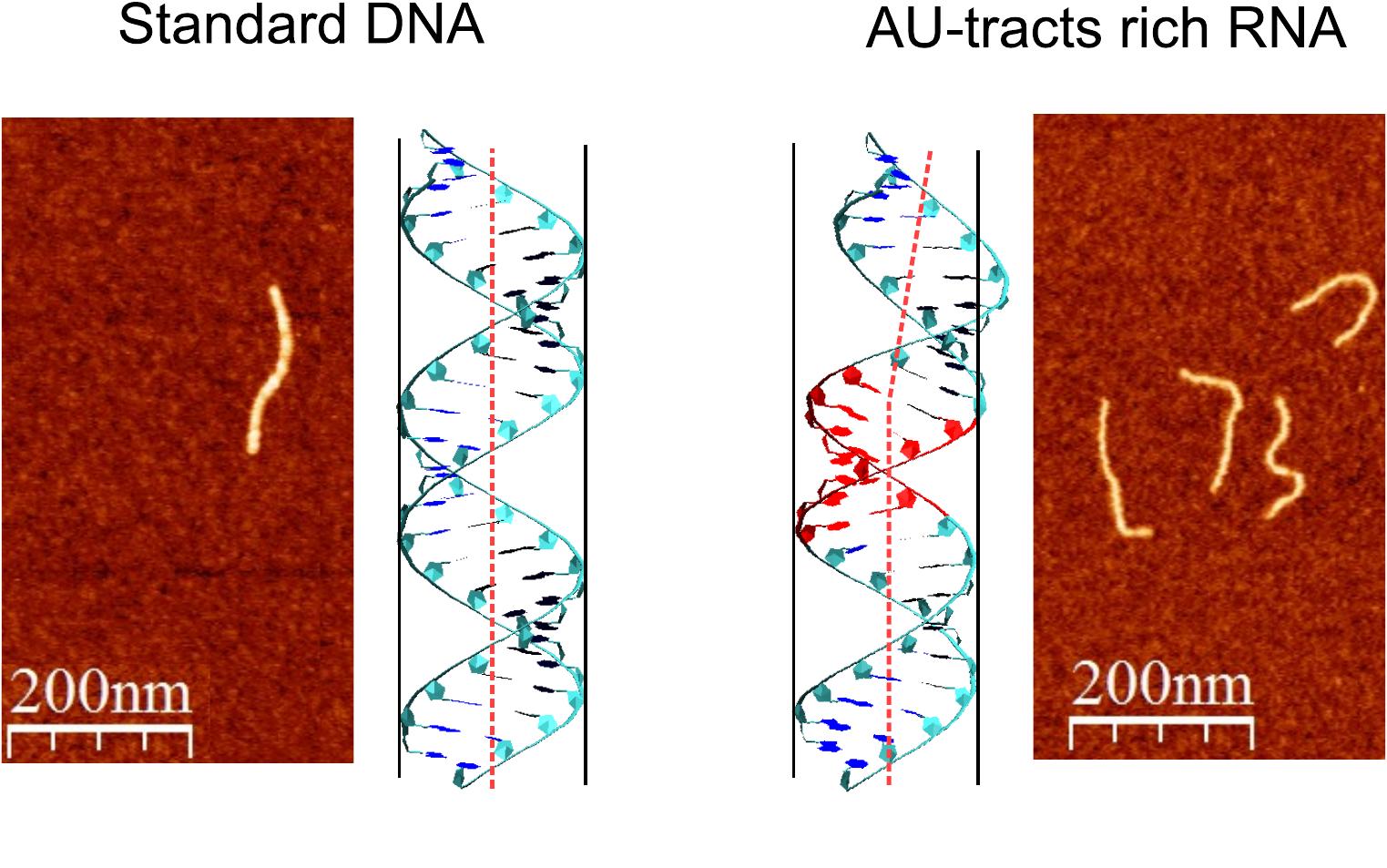
A. Marín, C. Pastrana, R. Bocanegra, A. Martín, J. G. Vilhena, R. Perez, B. Ibarra, C. Aicart and F. Moreno. “Understanding the paradoxical mechanical response of in-phase A-tracts at different force regimes“. Nucleic Acids Research 48 (9), 5024 (2020).
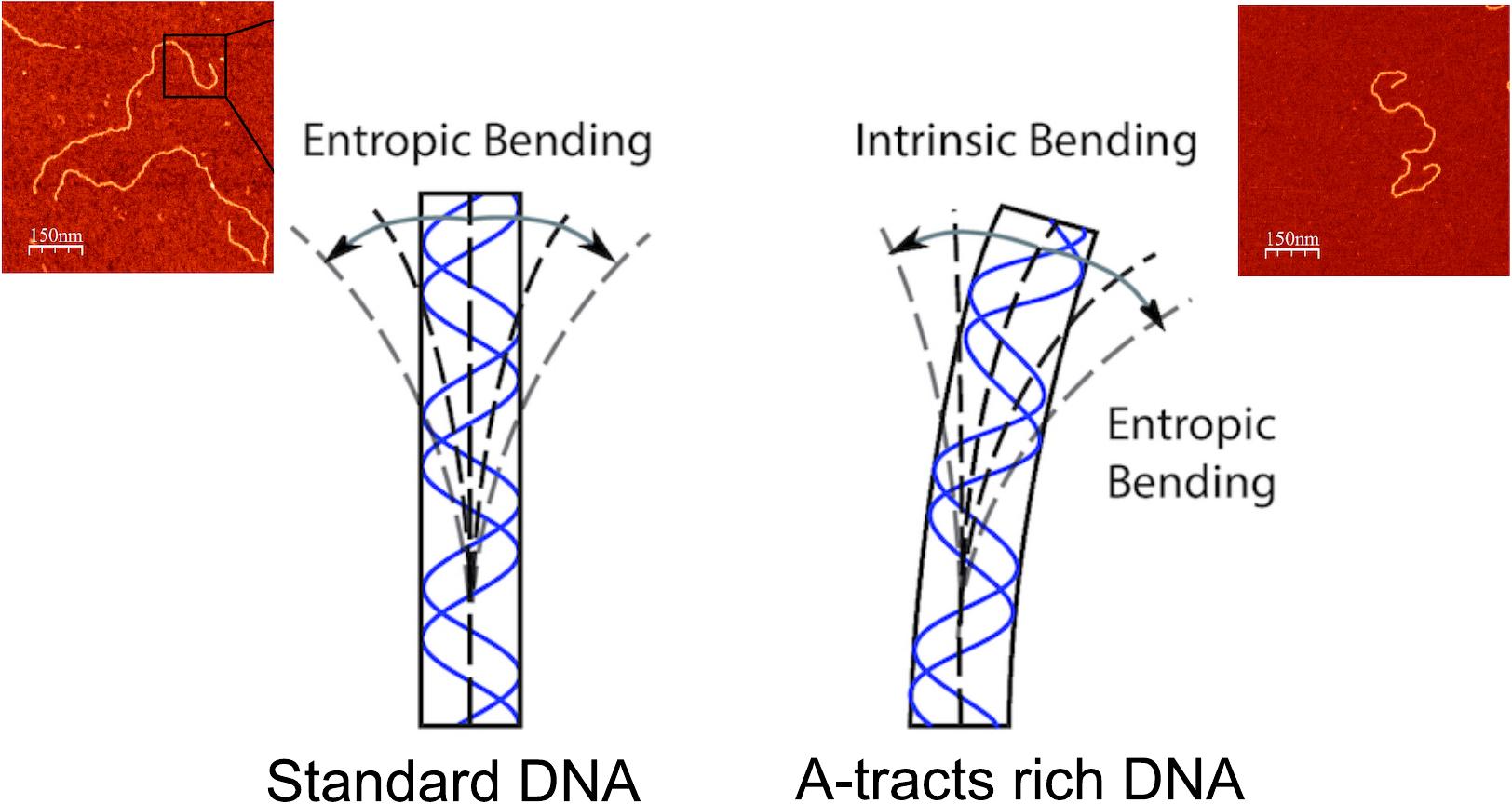
A. Marín, J. G. Vilhena, F. Moreno and R. Pérez. “DNA crookedness regulates DNA mechanical properties at short length scales”. Physical Review Letters 122 (4), 048102 (2019) -– featured in Nature Reviews Physics.
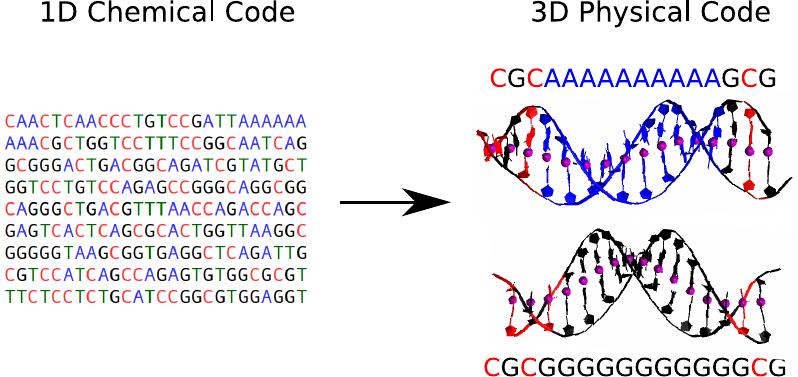
A. Marín, J. G. Vilhena, F. Moreno and R. Pérez. “Sequence-dependent mechanical properties of double-stranded RNA“. Nanoscale 11, 21471 (2019).
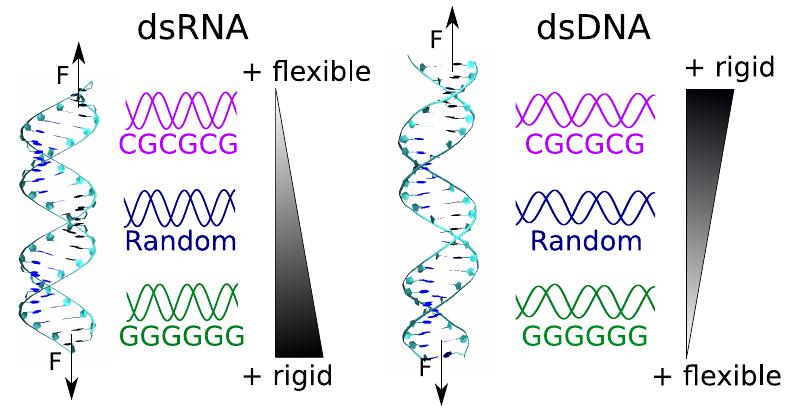
A. Marín, J. G. Vilhena, R. Pérez and F. Moreno. “Understanding the mechanical response of double-stranded DNA and RNA under constant stretching forces using all-atom MD“. Proceedings of the National Academy of Sciences USA 114 (27), 7049 (2017).